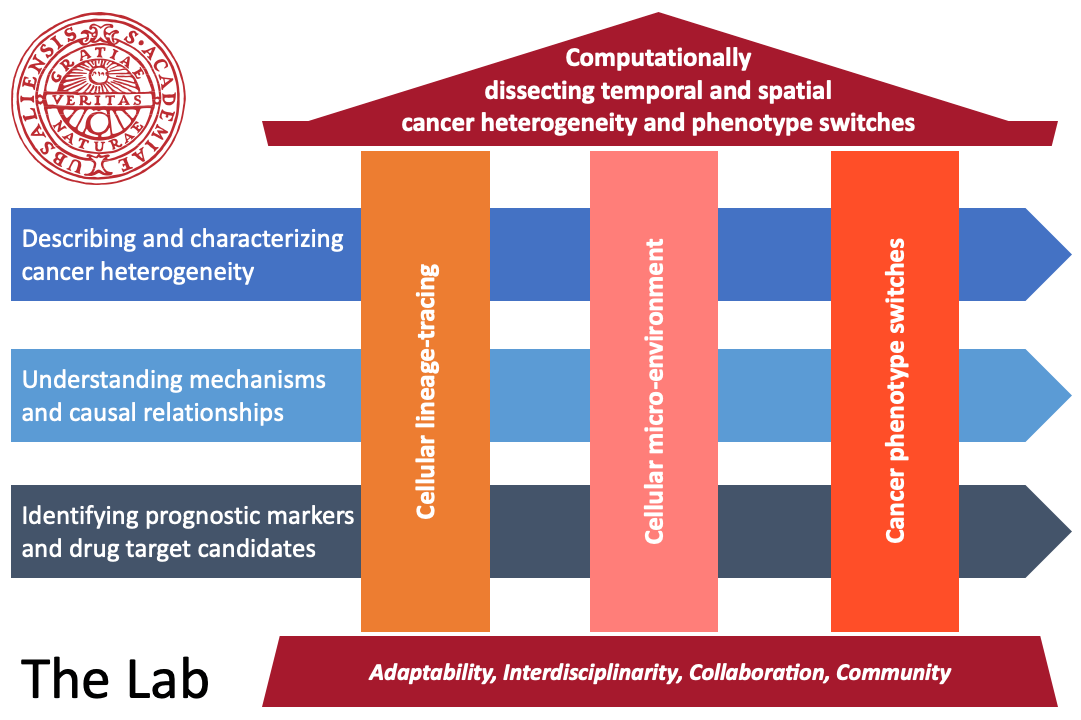
We develop and apply innovative computational tools to infer and integrate complex cell features (e.g., cellular ancestries and micro-environment) to dissect intra-tumor cancer heterogeneity, especially phenotype switches (e.g., acquisition of therapy resistance and tissue invasion). Our aim is to identify and characterize markers for disease progression, molecular phenotypes that can guide treatment and novel therapeutic targets to improve patient care through precision medicine. While working purely computationally, we apply an interdisciplinary approach bringing together cutting-edge molecular biology, technology and algorithms – together with our collaborators and partners in academia and industry.
Group members
TBD (You? Get in touch!)
