My group has three major lines of research:
1) To clarify the evolution of the mechanisms for long-term memory.
The molecular machinery for memory formation constitutes one of the most challenging questions for neuroscientists to resolve. Such complex mechanisms can only be understood by studying their components. Our approach is to investigate how and when the genes and their proteins originated that are involved in these processes. We already know that most of them arose at the origin of the vertebrates more than 500 million years ago. We use the zebrafish as a model species to investigate the localization and functions of the memory proteins.
2) To resolve the evolution of important gene families in vertebrates, particularly gene families expressed in the nervous system and in the endocrine system.
The purpose is to discover at which point new functions have arisen and how functions have changed during evolution. We are primarily investigating gene families that include neuropeptides, G-protein-coupled receptors, ion channels, and genes involved in vision.
3) To investigate functionally the subtle properties of G protein-coupled receptors that make them bind and respond to certain ligands and turn down others.
We are focusing on adrenergic and dopaminergic GPCRs because their native ligands are highly similar, yet the 11 adrenergic and 9 dopaminergic receptors in vertebrates (9 and 5, respectively, in mammals) usually have clear ligand preferences. We explore these therapeutically important differences by using evolutionary comparisons, structural modelling, mutagenesis, functional expression and pharmacological characterization (binding and signal transduction). This will increase our understanding of the receptor preferences of the many synthetic agonists and antagonists that are clinically important. Ultimately, we plan to design novel ligands as well as modified receptors to improve pharmacological therapy.
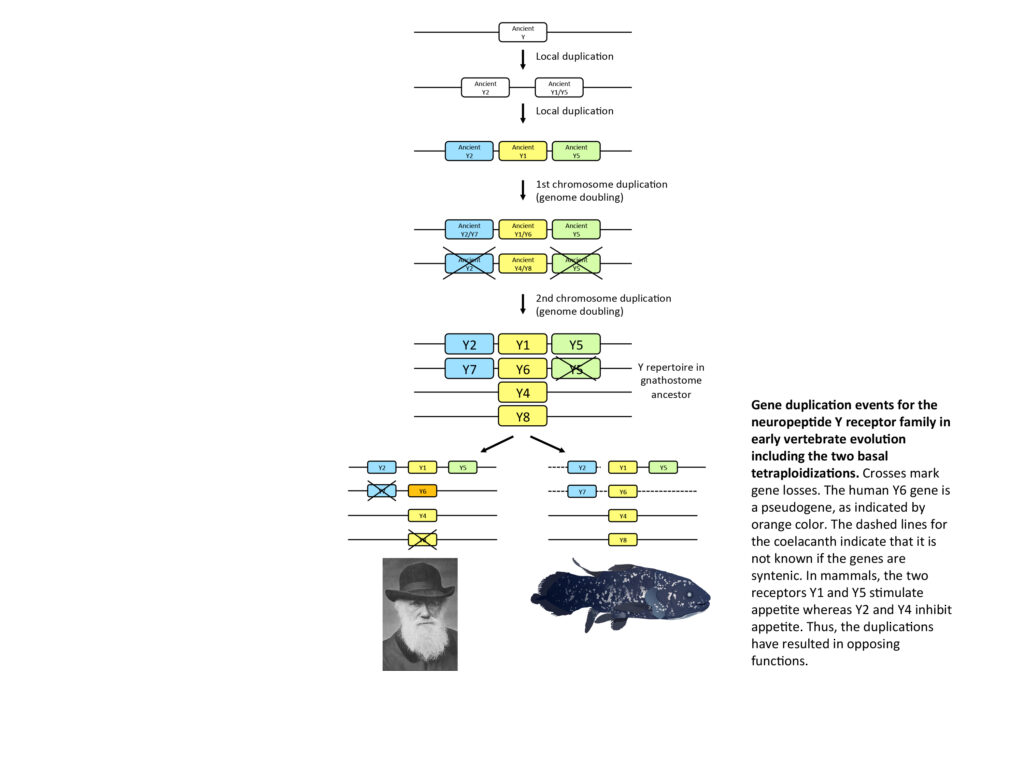
Many hundreds, perhaps thousands, of vertebrate gene families are now known to have expanded in two dramatic events that took place approximately 500 million years ago, namely two genome doublings or tetraploidizations (called 2R for two rounds of genome duplication). In addition, a third tetraploidization (3R) took place in the ancestor of teleost fishes. These events explain a great deal of the complexity of presently living vertebrates, and also explain functional overlap for members of many gene families. We are using a combination of phylogenetic sequence analyses and chromosome comparisons across species, to distinguish gene duplication events in gene families of special functional interest. This approach is very useful to identify corresponding genes (orthologues) in different species for comparisons of functions. The results have important implications for our ability to understand how functions arise, change, and occasionally even disappear during evolution. Among the gene families that we have studied, or are presently studying are the peptides and receptors in the neuropeptide Y family, the opioid peptides (enkephalins etc.) and their receptors, growth hormone and prolactin and their receptors, oxytocin-vasopressin receptors, somatostatin receptors, voltage-gated sodium and calcium channels, and receptors for the main neurotransmitters glutamate, acetylcholine, GABA, noradrenalin and serotonin.
Group members
Christina Bergqvist, research engineer
Helen Haines, PhD student
David Lagman, PhD, researcher
Jasna Pruner, PhD student
Yanying Li, PhD, postdoc
