About the PULSE program
PULSE builds upon and further leverages the unique SciLifeLab community, fostering excellence in biology-, technology-, and data-driven research. The program emphasises key research areas within molecular life science, and offers a well-structured, supportive environment with exceptional postdoc conditions to ensure excellent quality of individual projects and the overall program. PULSE adheres to principles for Open and FAIR data, and a Green Charter, ensuring maximum impact and contributing to long-term sustainability. To reach its full potential, PULSE will take the gender dimension, diversity aspects and unconscious bias into account throughout all parts of the project.
Skip to
Summary
Research areas
Key functions and constellations
Employment conditions for postdocs
Ethics issues and aspects
FAIR, Open Science and Green charter
Access to cutting edge research infrastructure
Intellectual Property Rights (IPR)
Additional information

Summary
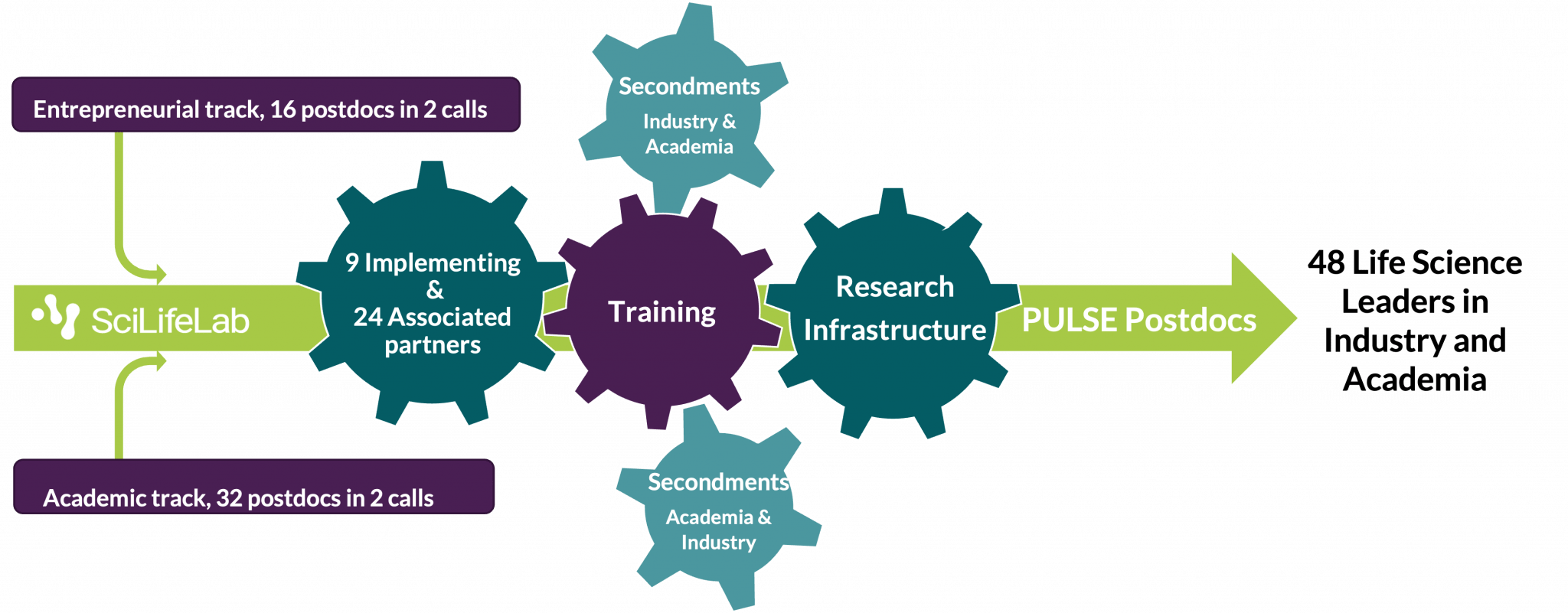
The purpose of the program is to strengthen the research network in Sweden and abroad in the long term by increasing the recruitment of international, highly qualified postdoc candidates in Life Science, and through mobility between universities, research infrastructures, research institutes and companies within the program.
SciLifeLab invited representatives from the Universities of Sweden, as well as representatives from the Life Science industry to participate in the design of the program. The resulting international consortium will ensure excellence and the implementation of the program in the best possible way.
The program has 9 university implementing partners, and 24 associated partners representing research infrastructures, research institutes and companies, that will contribute to postdoc training and host secondments.
PULSE will offer a unique setup, enabling candidates to choose between academic (32 positions) or entrepreneurial (16 positions) tracks. There will be 48 postdocs, recruited in two phases:
- Call 1 opening on January 7, 2025: 16 positions on the academic track and 8 positions on the entrepreneurial track
- Call 2 in January 2026: 16 positions on the academic track and 8 positions on the entrepreneurial track
Research areas
PULSE postdoc candidates apply with an individually-designed project, in one of two tracks within molecular life science.
Academic track
PULSE academic track postdocs will receive training in research groups seeking answers to complex and demanding questions of fundamental biological importance. Projects will be carried out in close proximity to the interdisciplinary analytic platforms at SciLifeLab equipped with cutting-edge technologies.
A common feature of ongoing projects is that they rely on methodologies and technologies that generate high density data.
PULSE postdocs will have the option to carry out projects within four major areas of research:
- Cell & Molecular Biology
- Precision Medicine & Diagnostics
- Epidemiology & Infection biology
- Evolution & Biodiversity.
Focus area descriptions
This area integrates the disciplines of genetics and biochemistry with the ambition to fully understand how cells, the fundamental unit of life, function. Progress is made by analyzing the molecular components in time and space, from single molecules to native tissue environments at high resolution. This research area is rapidly evolving with the development and application of novel high-density data driven methods, often relying on machine learning, artificial intelligence, or other computational techniques to analyze, integrate and interpret large sets of imaging data. The ultimate goal is to create predictive models for core functions of cells and tissues.
At SciLifeLab, research in this area is aligned with the EU definition of precision medicine, ie, “characterization of individuals’ phenotypes and genotypes to tailor therapeutic strategies, assess disease predisposition, and enable timely, targeted prevention.” Accordingly, here the aim is to integrate molecular and clinical data to identify biomarkers for disease risk and diagnosis, evaluate drug responses, and support health monitoring. Research projects are expected to contribute with strong capabilities inmachine learning, AI and other computational tools. Possibilities exist to mine strong assets in Sweden, such as molecular data (e.g. omics), imaging, electronic healthcarerecords, longitudinal patient and population registries, biobanks and digital monitoring data.
This subject area concerns research using large experimental, clinical, or pathogen surveillance data in innovative ways to transform our understanding of pathogens, their interactions with hosts and the environment, and how they are transmitted through populations. The aim is to meticulously analyze pathogen-host model systems, exploiting multidimensional and genome-scale experimental data that are now routinely attainable, and extending to population-scale genetic, clinical, or public health data from pathogen surveillance efforts and biobanks. The ultimate goal is to enhance the ability to anticipate, prevent, and respond to infectious disease outbreaks, ensuring a proactive approach to safeguarding public health.
Projects within this area address life from a trans-disciplinary perspective, encompassing molecules and cells to ecosystems. Researchers are leveraging the substantial data streams generatedby high-throughput sequencing of genomes and biomes,continuous recording of video and audio in the wild, high-throughput imaging of biological specimens, and large-scale remote monitoring of organisms and habitats. Novel insights are likely to emerge by exploiting the development and applicationof novel methods relying on machine learning, artificial intelligence, or other computational techniques, enhancing our understanding of population genetics, adaptation, speciation, convergent evolution, and functional constraints. A deeper understanding of the complex interactions within ecosystems and their implications for planetary health is needed to find solutions to impede the ongoing loss of biodiversity.
Entrepreneurial track
Development of disruptive technologies within the areas below to facilitate target-based drug discovery of novel therapeutic agents:
- Machine Learning/AI
- Therapeutic Oligonucleotides
- Display and Selection technologies
- Proximity Inducing Agents
Project ideas expected to advance from technical readiness level (TRL) 1-2 to TRL 5-6 in close collaboration with the SciLifeLab Drug Discovery and Development platform (SciLifeLab DDD), the associated academic innovation support system and the associated industrial partner. PULSE postdocs entering the entrepreneurial track will receive extensive and individual training as scientific leaders and entrepreneurs: both advanced academic training, and entrepreneurship training, including the steps required to create new companies focused on drug discovery and discovery. The Swedish teacher’s exemption gives the PULSE postdoc employed by a University, the right to their patentable inventions
Main PI/SciLifeLab Group Leader host is a SciLifeLab DDD Head of Unit, a Platform Scientific Director at SciLifeLab DDD or, a PI with complementary expertise and know how.
Focus area descriptions
This is an exceptional opportunity to conduct research at the intersection of artificial intelligence (AI)/Machine Learning (ML), data science, and drug discovery. The fellow will lead the design, development, and implementation of novel AI/ML methods for preclinical drug discovery of small molecules, proteins, or oligonucleotides, focusing on developing and applying AI/ML methods in hit-to-lead optimization projects for any of these drug modalities.
Key project areas include, but are not limited to, the development of AI/ML tools, web-based workflows, and novel approaches in deep learning, de novo design and generative AI for drug discovery. Candidates are encouraged to apply AI/ML methodologies across preclinical drug discovery project based on small molecules, proteins, oligonucleotides, or emerging “new modalities”. Examples of areas of research could be: novel molecular representations and generation, chemical space navigation, ultra-large virtual screening, multi-parameter optimization, ligandability assessment, protein design, RNA and oligonucleotide structure prediction, prediction of on/off-target oligonucleotide binding and effect, computational characterization of enzymatic RNA cleavage, antibody developability, and de novo design. We also welcome proposals from emerging fields in drug discovery, including explainable AI, and integrating ‘first-principles’ with AI/ML (e.g., quantum machine learning approaches).
Examples of challenges in this area:
•There is very limited access to high-quality, standardized datasets, combined with imbalanced data (e.g., active vs. inactive compounds) which hampers model training and generalizability. How to access these data sets?
•Many models, particularly deep learning-based ones, suffer from a lack of interpretability, making it difficult to validate predictions and gain insights into molecular mechanisms. Methods to solve this problem?
•Scalability is another barrier, as handling large datasets or running complex simulations requires significant computational resources. How to speed up things?
•The interdisciplinary knowledge gap between AI researchers and domain experts in chemistry and biology slowsprogress, as effective collaboration requires bridging expertise across these fields. Can these barriers be torned down?
The successful candidate will join a vibrant, multidisciplinary research environment, collaborating with bioinformaticians, structural biologists, chemists, and drug discovery experts.
Through the OligoNova Hub units in Gothenburg, SciLifeLab-DDD offers state-of-the-art capabilities for oligonucleotide-based drug discovery, including but not limited to antisense and siRNA-based therapeutic modalities, as well as modalities that conjugate oligonucleotides to SciLifeLab-DDD’s other modalities such as proteins and small molecular ligands. In-house capacity for bioinformatics and AI/ML-guided oligo design, oligonucleotide synthesis, automated and high-throughput analyses of cellular effects through qPCR, NGS, mass spectrometry, imaging, and other techniques is complemented with advanced proteomics, NMR, and additional imaging modalities including Nano-SIMS within the local core facilities organization and other collaborators.
New technologies to design and analyse therapeutic oligonucleotides to support the development of optimally efficacious and safe therapeutic agents are continuously developed and applied within the focus area. Current examples include the development of a modular reporter assay system to evaluate target knockdown, new computational techniques for nucleic acid structure prediction, and biophysical technologies for the characterization of oligonucleotide binding interactions.
Of great interest are new technologies and approaches that could streamline any aspect of the discovery process, provide new molecular and/or mechanistic insight into the action and safety of therapeutic oligonucleotides, provide new means of measuring oligonucleotide distribution, exposure, and effects in vitro and in vivo, or enable the design of novel tissue- and cell type-selective oligonucleotide delivery strategies. In addition to experimentally oriented projects, AI/ML-approaches or other computational techniques could be integrated project components.
Examples of challenges in this area:
•New methods to characterize and prioritize therapeutic oligonucleotides in large libraries with regard to efficacy and/or safety
•New methods to measure tissue- and (sub)cellular distribution of oligonucleotide therapeutics
•New strategies for enhancing delivery of oligonucleotide therapeutics to specific tissues and cell types through conjugation to targeting molecules, and new methodologies to identify and characterize such targeting strategies
•Methods for better prediction of stability and tolerability/safety of therapeutic oligonucleotides
Contact: par.matsson@gu.se
Within the SciLifeLab Display and Selection Technologies focus area we have access to state-of-the-art platforms for discovery of therapeutic antibodies, small molecules and oligonucleotides. These include e.g. phage display libraries as source of human antibodies, DNA-encoded chemical libraries (DELs) for small molecules, automated systems to facilitate high-throughput screening and instruments to study binding of therapeutic candidates to proteins and cells. Through close collaborations with other SciLifeLab units, local university networks and CROs additional instruments can be accessed e.g. mass spectrometry, DNA sequencing (Sanger, NGS, long-read) and various imaging techniques.
Technology development is continuously ongoing within the focus area. Currently a single-domain antibody library in phage display format as well as new DELs are under construction, antibody tools for targeted protein degradation and intracellular delivery are developed, new reporter systems for screening of oligonucleotides are set up and long-read sequencing is explored as a complement to traditional antibody screening.
We welcome technology development proposals improving any step of the early drug discovery process. Areas of interest could include e.g. novel selection and screening strategies, alternative display technologies, new therapeutic antibody formats, strategies for optimization of binding affinity and strategies for improved developability. AI-driven approaches and NGS/long-read sequencing could be integrated elements in such development work.
Examples of challenges in this area:
•Presently, recombinant protein displayed on beads are used for DEL screening, Can a technology be developed that allows for in situ selection from large DELs in human cells or bacteria?
•Can NGS-based strategies be developed to bypass needs for screening the output from selections in phage display libraries?
•Engineering of candidate drug antibodies for preclinical development is circumstantial. Can new strategies be developed to make human antibody maturation and the optimization of developability aspects more efficacious?
•Novel technologies and vectors that generate a better therapeutic window for CAR applications are in demand. How can this be accomplished?
•Development of novel display/array formats for antibody/peptide screening and development
Contact: mikael.mattsson@immun.lth.se
Proximity-Inducing Agents are an emerging approach of therapeutics that leverage the ability to bring two or more biomolecules into proximity to induce specific biological effects. These agents are based on technologies such as bi-specific antibodies, antibody-drug conjugates, or various small molecule approaches for intracellular re-wiring of protein localization. Emerging technologies for Targeted Protein Degradation (TPD), such as PROTACs (Proteolysis Targeting Chimeras) and AbTACs (antibody-based targeted protein degradation agents), show promise in precisely targeting and degrading disease-causing proteins, potentially offering new treatment avenues that conventional drugs may not address.
We offer exceptional opportunities to conduct research that will shape the technological advancement of proximity inducing therapeutics based on modalities that is compatible with areas within scope for the SciLifeLab DDD platform, i.e. small molecules, proteins, oligonucleotides, or new modalities. Cell-based therapies are out-of-scope.
Depending on the candidate’s background and interests we support proposals related, but not limited, to: small molecule degraders, antibody-based degraders for extra-cellular or membrane proteins, targeted delivery of payloads (oligonucleotides, small molecules, radioligands, degraders), bi-specific antibodies, etc. Currently, we have toolboxes, assays, and methodology for small molecule- or antibody-based degraders and targeted delivery available. We offer industry standard facilities and infrastructures for e.g. organic synthesis – including oligonucleotides, biochemical and cellular assays, biophysics, ADME, protein expression, phage-display selections, computational and machine learning capabilities, etc. SciLifeLab offers opportunities to access additional world-class infrastructures.
Examples of challenges in this area:
•New methods that enable the identification of bifunctional compounds and molecular glues that act on pre-selected presenter and target proteins, e.g. by comparing barcode enrichment after screening DNA-encoded libraries in the presence or absence of presenter proteins
•Novel antibody formats with applications for targeted protein degradation
•Innovative technologies that identifies drug-like molecules with high affinity to unstructured proteins
Contact: Per.Arvidsson@scilifelab.se
Proposed projects at the intersection of, or that combine any of these areas, are welcome.
Key functions and constellations
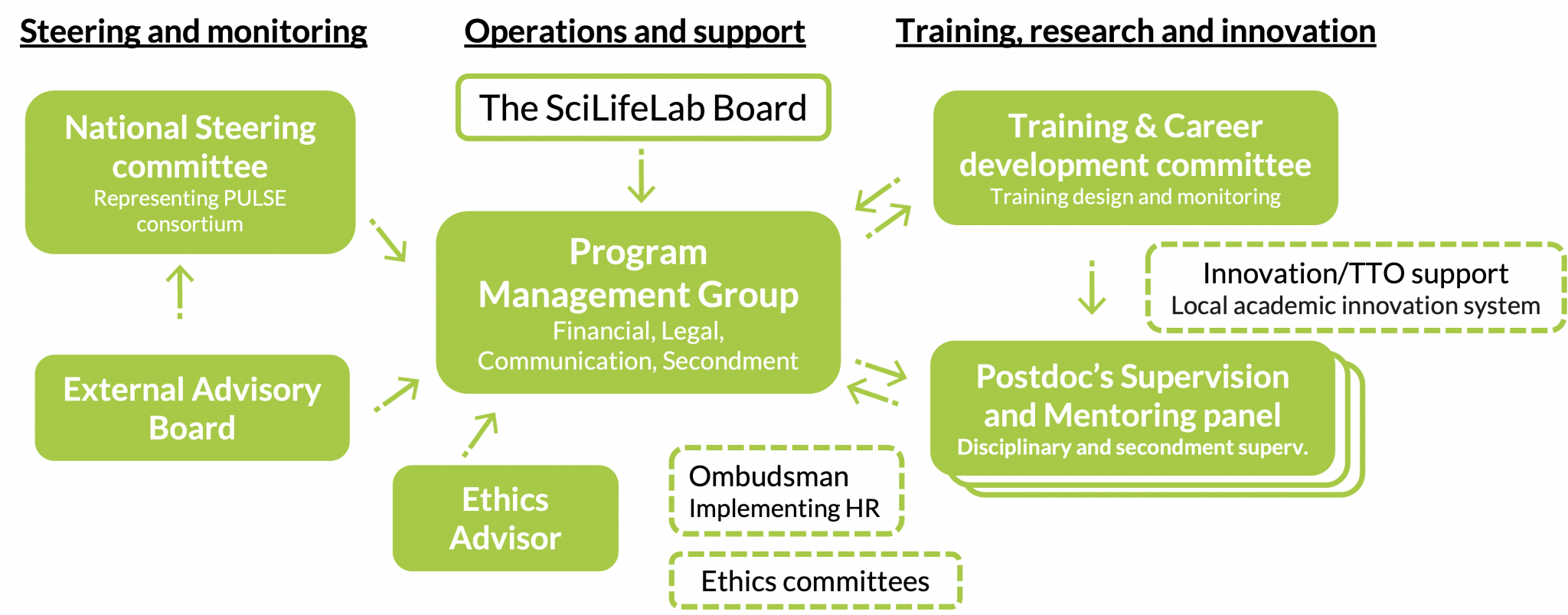
PULSE builds upon and integrates with SciLifeLab’s well-established organisational structure, with functions for Steering and monitoring, Operations and support and Training, research and innovation. PULSE postdocs will be hosted by one of the Implementing partners.
The SciLifeLab community recognizes the importance of a healthy and safe work environment for productivity and success. The SciLifeLab Code of Conduct guides responsible conduct in research infrastructure services, research, and training, ensuring a respectful and healthy working environment, and a positive impact on society. The code covers ethical dimensions vital to the community’s sustainable growth, promoting responsible conduct and ethical practices.
The National Steering Committee (PULSE-NSC) makes decisions regarding postdocs selection, budget & any issues during the program. PULSE-NSC have the role of Scientific and Impact committee, assure national focus and that postdocs are embedded in SciLifeLab and local environments.
The External Advisory Board will give strategic advice on PULSE training.
The PULSE Program Management Group (PMG) is responsible for overall programme management, risk assessment and management.
An Ethics Advisor will be tasked to identify and follow-up ethics aspects of the overall project.
PMG members:
- Mia Phillipson, SciLifeLab Co-Director, Director of SciLifeLab PULSE
- Per Ljungdahl, Campus Solna Director, Co-Director of SciLifeLab PULSE (academic track)
- Kristian Sandberg, Platform co-Director DDD, Co-Director of SciLifeLab PULSE (entrepreneurial track)
- Jessica Lindvall, SciLifeLab Training Hub Lead, Director of SciLifeLab PULSE Training
- Disa Hammarlöf, Program Manager SciLifeLab PULSE
- Maria Bäckström, Program Coordinator SciLifeLab PULSE
The PULSE Supervision & Mentoring Panels with representation from both the Implementing and Associated partners, along with the Training and Career Development Committee, will ensure excellent individual training and career development support, maximising the success of each PULSE postdoc. The local academic support system at the Implementing partners will support in exploitation of relevant PULSE projects, and contribute in entrepreneurship training.
PULSE supervision & mentoring panel
Each PULSE postdoc will have a SciLifeLab Group Leader as main supervisor at one of the Implementing host institutions. SciLifeLab PIs have strong scientific track records, and offer an exceptional scientific milieu in close proximity to the SciLifeLab research infrastructure. The supervision panel will also hold two co-supervisors with complementary expertise, eg from other disciplines and/or sectors, from either of the PULSE Implementing or Associated partners, and can be affiliated with the secondment host. For the entrepreneurial track, one of the co-supervisors is a DDD project leader. Feasibility of the project is the responsibility of the supervisors (access to infrastructure, ethical approvals needed etc).
Each PULSE postdoc will also be assigned a mentor from the PULSE consortium, who will actively support and promote the postdoc’s career development.
Training and Career Development Committee
The Training and Career Development Committee (TCDC) will design and monitor the PULSE program training and the postdocs’ training progress based their individual Program Career Development Plan (PCDP).
Working with their supervisors and the TCDC, each PULSE postdoc will set up a PCDP. The PCDP will be updated annually to ensure that the training is tailor made for each postdoc. Once a PCDP has been developed, PULSE postdocs should feel empowered and responsible for their career development. Read more about the PULSE training program.
Employment conditions for postdocs
PULSE postdocs will be offered an employment with a competitive, minimum starting gross salary of 38,000 SEK/month (est. 3326 EUR/month). This equals to net salary of 30,000 SEK/month (est. 2605 EUR/month). In Sweden, additional social benefits are provided through the Swedish social security system, including eg parental leave & subsidised childcare and sick-leave allowance. The Implementing partners offer staff benefits to promote health and fitness, support for researchers with disabilities and researchers at risk.
Relocating and living in Sweden
All PULSE Implementing partners‘ hosting institutions provide free personalised assistance in matters related to relocation to Sweden, e.g. visa applications, setting up bank accounts, enrollment in Swedish administrative and social security systems, health care, and housing (with support from local offices specialising in international recruitment of talent). Find out more about relocating to and living in Sweden by contacting the EURAXESS Service Centre (ESC) of interest. Free tuition in Swedish is provided by SFI Swedish for immigrants.
Learn more about the cities where the program hosts are placed: Gothenburg, Lund, Linköping, Stockholm, Umeå, Uppsala and Örebro.

Ethics issues and aspects
During application
All applicants must submit an Ethics self-assessment in digital form (if ethical approval is needed for the proposed project, this should be stated in application)
During the application review process
If a selected project needs ethical approval, the PI main host is responsible to move forward with the process and apply for ethical approval. SciLifeLab PULSE requires that the ethical approval is in place before starting the project.
For activities undertaken in non-EU countries and that raise ethics issues, a confirmation that the research is legal in at least one EU Member state will be required.
An independent Ethics Advisor will identify and follow-up ethics aspects during the recruitment process.
Upon recruitment, any ethical issues related to personnel administration are handled by the relevant HR department.
During the program
An independent Ethics Advisor will identify and follow-up ethics aspects through-out the program.
The Training and Career development committee, consisting ofTraining Director, Training Coordinator and local training representatives, will handle ethics issues in relation to supervision, together with the local ethics experts at the host institution.
If conflicts arise involving postdocs, supervisors, and/or committee members, the PULSE Management Group will synchronize efforts to resolve them through the relevant implementing partner HR department (the primary responsible party) and the PULSE Ethics Advisor.
Fair, Open Science and Green charter
The PULSE program emphasises FAIR (Findable, Accessible, Interoperable, Reusable) principles, Open Science, and the Green Charter to promote transparent, reproducible research while reducing the environmental impact of scientific activities. These frameworks ensure that data and findings are widely accessible and reusable, fostering global collaboration and innovation. Moreover, integrating sustainability practices helps align research with broader environmental goals, addressing both scientific and ecological responsibilities.
Data generated in PULSE projects will be managed in line with the FAIR principles (Findable, Accessible, Interoperable, Reusable). Data will be made available through open platforms or transparent controlled access procedures when appropriate, with tools and support through SciLifeLab Data Centre and NBIS.
Open Science practices are central to PULSE. Sweden has national guidelines for Open Science and all Swedish universities operate under the Open Science Plan that is based on EU and UNESCO guidelines. The PULSE Implementing partners all have Open Science Strategies and resources, and PULSE will rigorously train researchers in the handling of big data and Open Science.
PULSE will adhere to the principles of the MSCA Green Charter at multiple levels. Institution level. All participating ImplPrtns have ‘green guidelines’ and institutions are expected to apply them on a day-to-day basis, e.g. in terms of reduce, reuse and recycle material both for daily use and for specific materials and chemicals. Postdocs will be briefed upon their institutional green practices at the start of their employment. Project level. Postdocs will have dedicated training on the EU Green Charter and the MSCA green charter with updates as necessary. Two PULSE Implementing partners are listed in the MSCA Green Charter: The Beneficiary KTH Sustainability Office, dedicated to the integration of environment and sustainability in education, research and collaboration, and the Lund University Centre for SUstainable Studies (LUCSUS), and will serving as a paradigm to inspire all participating PULSE partners to take corresponding steps of their own.
Access to cutting edge research infrastructure
SciLifeLab is one of the largest European Research Infrastructures in Life Science. During the entire fellowship PULSE postdocs will have access to the SciLifeLab infrastructure, which provides access to 10 cutting-edge technology platforms in molecular biosciences: Bioinformatics, Genomics, Clinical Genomics, Clinical Proteomics and Immunology, Metabolomics, Spatial Biology, Cellular and Molecular Imaging, Integrated Structural Biology, Chemical Biology and Genome Engineering, and Drug Discovery and Development. Access to this technology and expertise will contribute to the postdocs obtaining advanced research skills.

Intellectual property rights (IPR)
Welcome to Sweden and the teacher’s exemption – you own your research results!
Please note, the text below is not a legal recommendation. You must ensure the details regarding the teacher’s exemption for you at your host university. This text merely aims to introduce how academic intellectual property rights are handled in a Swedish context.
As a researcher in Sweden, you will have a unique opportunity as well as responsibility when it comes to the ownership of your research results. The Act on the Right to Employees Inventions (1949:345; the “Inventors Act”), dating back from 1949, in principle states that you as a researcher at a university in Sweden own the patentable inventions originating from your research. This is referred to as “teacher’s exemption” since it exempts the universities from obtaining the intellectual property rights from the researcher/employee which is typically the case for private companies. It is sometimes referred to as the “professor’s privilege” which is misleading since the scope is broader than only professors. Each university may also have interpretations regarding both which personnel are included in the teacher’s exemption beyond researchers, as well as what rights that belong to the researcher. Furthermore, the universities reserve the right to use the results without additional compensation for the ordinary academic operations at the university.
The teacher’s exemption influences how the university engages in external collaborations and needs to be regulated in agreements with all parties involved including the researcher to ensure that all stakeholders are in agreement.
As a researcher with a program accepted at the Drug Discovery and Development platform (DDD) it is worth mentioning that intellectual property rights stay with you as a researcher. The details will be specified in an agreement between the researcher, university and the host universities for DDD.
Each university has different support functions for researchers in regard to transforming and developing research ideas towards services or products for the benefit of society. It is highly recommended to contact support functions at your host university for additional information and support.
The subject will be further explored in the SciLifeLab PULSE training activities.
Additional information
Want to distribute information about PULSE in PDF format? Download the PULSE flyer!

N.B. Funded by the European Union. Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or the European Research Executive Agency. Neither the European Union nor the granting authority can be held responsible for them.