New study sheds light on DNA movements during nucleosome remodeling.
Our genetic material, the DNA, is packaged into a tightly condensed structure referred to as chromatin. The chromatin is in turn made up of nucleosomes which all contain a stretch of DNA wrapped around a cylinder-shaped core consisting of histone proteins.
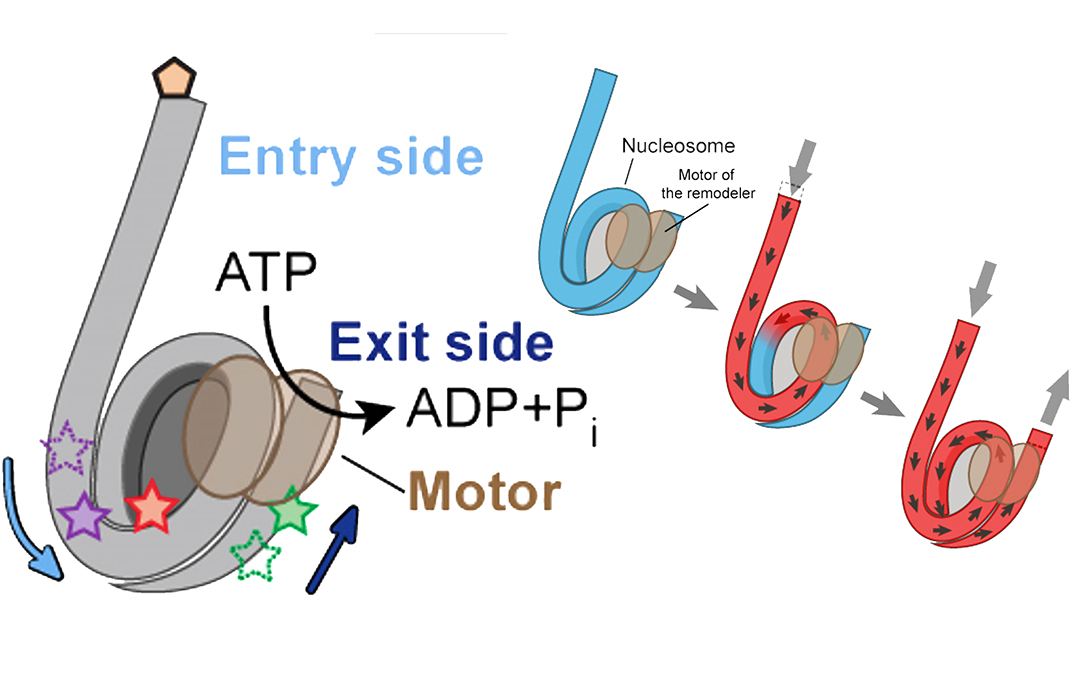
Inside the nucleosome, the DNA is largely shielded from interactions with proteins, which means that the structure has to be remodeled in order for the cell to obtain access to the DNA. One way this can be achieved is through the action of protein machines called chromatin remodelers. These remodelers can “slide” nucleosomes along the DNA, leaving certain portions of it exposed.
In order to achieve nucleosome sliding, the remodeler needs to feed DNA onto one side of the nucleosome and release it on the other side, thereby breaking and re-forming DNA-histone interactions as the nucleosome moves along the DNA – a mechanism that is poorly understood.
Due to the fact that remodelers operating on different nucleosomes are not synchronized, it is not sufficient to observe an average signal from a collective of nucleosomes. Observations therefore have to be made at the level of an individual nucleosome and, in order to understand how movements are coordinated, simultaneously on both of its sides.
In a new study, published in Nature Communications, led by Sebastian Deindl (SciLifeLab/Uppsala University) and Greg Bowman (Johns Hopkins University), researchers, including Anton Sabantsev (SciLifeLab/Uppsala University) and Rob Levendosky (Johns Hopkins University), investigated the mechanism of nucleosome remodeling by using a new powerful three-color single-molecule FRET approach – a technique that can measure two distances within a single molecule simultaneously – and combining it with high-resolution crosslinking data to determine the register of DNA on the nucleosome during remodeling.
The researchers succeeded in observing how the real-time DNA movements on both sides of the nucleosome are coordinated. The remodeling occurred in a sequential manner with DNA first being pulled onto the nucleosome but not shifted off the other side. Instead, additional DNA was stored on the nucleosome, building up a spring-like tension until sufficient to push the extra DNA off the nucleosome on the other side.
This DNA buffering ability, which the researchers observed in real time, may represent a common property harnessed by other chromatin-interacting machinery.