Spatial Biology – The 2024 SciLifeLab Science Summit
October 1 @ 09:00 – 17:00 CEST
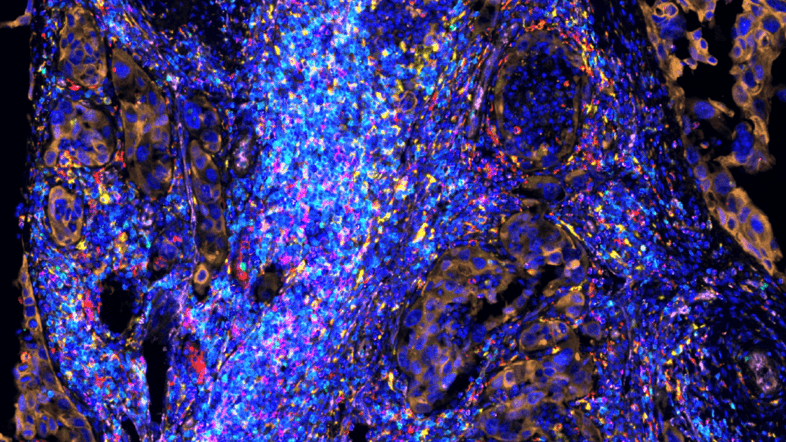
Spatial Biology is an emerging field driven by recent advances in Spatial Omics technologies. The aim of the summit is to present the spatial omics capabilities at SciLifeLab, and examples of uses of such methods to address various research areas in Life Science. A lot of pioneering work in Spatial Biology originates from researchers associated with SciLifeLab, and we hope to inspire more researchers to explore this approach and to join this growing spatial community.
The Annual Conference SciLifeLab Science Summit is a one-day symposium with a new topic each year. The topic for 2024 is Spatial Biology.
We warmly welcome international visitors to attend this free event. The conference also offers fantastic opportunities for networking and discussions with top academic researchers and industry leaders in spatial omics technology.
Welcome!
Mats Nilsson, Chair of the Scientific Committee, Charlotte Stadler, Per Andrén and Stefania Giacomello
Program
Posters
Poster Presentation Guidelines
Poster Format
The poster screens are 103 cm wide and 143 cm high. Please ensure your poster fits within these measurements.
Mount your poster
The venue will be open from 8:45 AM. Please proceed to the 6th floor and hang your poster at your designated number. A numbered poster list will be provided closer to the event date.
Poster sessions
Poster Session I: During the morning coffee break, from 10:55 AM to 11:35 AM, presenters of posters with odd numbers (1, 3, 5, 7…) should be present by their posters.
Poster Session II: During the afternoon break, from 3:15 PM to 3:55 PM, presenters of posters with even numbers (2, 4, 6, 8…) should be present by their posters.
Best Poster Award
All poster presenters should attend the Best Poster Award ceremony at the end of the conference, where the winners will be announced. The Scientific Committee will evaluate the posters throughout the day.
Dismount of posters
Please remove your poster immediately after the closing session at 5:00 PM. Poster screens will be collected at 5:30 PM, and any remaining posters will be discarded.
Poster List
Abstracts
Registration
For late registrations (after September 13), we cannot guarantee lunch, coffee, nor can we accommodate any allergies or dietary requirements. These will be subject to availability. We appreciate your understanding.
Questions? If you have any inquiries regarding the event or your registration, please contact events@scilifelab.se
The conference is free of charge; however, if you do not attend and fail to cancel, your department or organization will be charged a no-show fee of 500 SEK.
Important Notice Regarding Venue Regulations
Please be advised that for security reasons, Uppsala Konsert & Kongress (UKK) has a strict policy regarding bags and outside clothing.
Bags larger than A4 size and outside clothing (jackets, coats etc.) are not permitted inside the auditorium.
However, you are welcome to store these items in the venue’s cloakroom. Laptops can be brought into the auditorium in laptop sleeves.
We apologize for any inconvenience this may cause and appreciate your understanding and cooperation in adhering to the venue’s regulations.
Invited Speakers – abstract and bio sketch
Click on the arrow to read the speaker’s title, abstract, and bio.
Gonçalo Castelo-Branco, Karolinska Institutet, Sweden
Talk
Oligodendroglia in Development and Multiple Sclerosis: Insights from Single-Cell and Spatial Omics
Abstract
Oligodendroglia (OLG) mediate myelination of neurons, a process that allows efficient electrical impulse transmission in the central nervous system. An autoimmune response in multiple sclerosis (MS) leads to OLG cell death, loss of myelin and neuropathology. Using single cell transcriptomics, we have previously identified disease-specific OLG populations in the EAE mouse model of MS and in human MS brain archival tissue, characterized by the expression of immune genes.
By assessing chromatin accessibility and the transcriptome simultaneously at the single cell level at different stages of the disease course, we found that immune genes exhibit a primed chromatin state in mouse and human OLG in a non-disease context, compatible with rapid transitions to immune-competent states in MS. Moreover, we found dynamic and distinct transcriptomic and epigenomic responses of OLG subpopulations to the evolving environment in EAE mouse model of MS, which might modulate their response to regenerative therapeutic interventions in MS.
While single-cell genomics are powerful for investigating disease-specific cellular states, these methods involve isolating the tissue under study from its niche, leading to a loss of spatial information. Such information is essential for determining cell-to-cell communication in disease niches. We have applied in situ sequencing to investigate disease evolution in MS at a spatial level, both in the EAE mouse model of MS and in human post-mortem MS samples. We annotated disease neighborhoods during lesion evolution and found centrifugal propagation of active lesions. We demonstrated that disease-associated (DA)-glia arise independently of lesions and are dynamically induced and resolved over the disease course.
We have also applied dBIT-Seq, a ligation-based method for deterministic barcoding in tissue, to probe different histone modifications and chromatin accessibility in the mouse brain tissue sections, either in an unimodal or simultaneously with transcriptomics. This spatial epigenome–transcriptome co-profiling has allowed us to identify cellular lineage progression and epigenomic priming events that precede transcription during development with spatial resolution. We are currently applying these methods to disease paradigms in MS, to uncover how transitions to pathological cellular states occur at epigenomic and transcriptomic levels.
Bio
Gonçalo Castelo-Branco researches oligodendrocytes’ epigenetic states, focusing on how transcription factors, non-coding RNA, and chromatin enzymes drive cell changes. His work has potential for multiple sclerosis therapies.
Ron Heeren, Maastricht University, The Netherlands
Talk
Spatial biology and translation life sciences
Abstract
Modern molecular analytical technologies in the “omics” arena plays a crucial role in many scientific disciplines ranging from material sciences to clinical diagnostics. Technological advances have increased methodological sensitivity allowing researchers to acquire detailed molecular information of smaller and smaller samples. The biggest challenge is to put that concerted information in the context of the biological problem the samples originate from. This lecture describes how innovative mass spectrometry based molecular imaging technologies, have impacted translational clinical research and beyond. Or: how a mass spectrometer can be used as a sensitive and selective molecular microscope in modern spatial biology. Targeted and untargeted imaging technologies now offer new insights in the complexity that can be employed for systems medicine. Innovations in mass spectrometry based chemical microscopes have now firmly established themselves in translational molecular research. One key aspect of translational success is the ability to obtain this molecular information on thousands of molecules on a timescale relevant to translation. Single cells can be analyzed in great molecular detail and in the context of their native tissue. Combined this offers a true multi-omics approach that reveals contextual molecular complexity for systems medicine.
Bio
Prof. Dr. Ron M.A. Heeren obtained a PhD degree in technical physics in 1992 at the University of Amsterdam on plasma-surface interactions. He was the research group leader at FOM-AMOLF for macromolecular ion physics and biomolecular imaging mass spectrometry in the period 1995-2015. In 2001 he was appointed professor at the chemistry faculty of Utrecht University lecturing on the physical aspects of biomolecular mass spectrometry. In 2014 he was appointed as distinguished professor and Limburg Chair at the University of Maastricht. He is scientific director of M4I, the Maastricht MultiModal Molecular Imaging institute and heads the division of imaging MS. He is the vice-president of the international mass spectrometry foundation and has been active in many professional societies to advance mass spectrometric research, education and professionalization. His academic research interests are mass spectrometry based personalized medicine, translational molecular imaging research, high-throughput bioinformatics and the development and validation of new mass spectrometry based “omics” imaging techniques for the life sciences.
Arutha Kulasinghe, The University of Queensland, Australia
Talk
Uncoupling Pathways Involved in Immunotherapy Resistance: Insights from Deep Tissue Profiling
Abstract
Spatially resolved multi-omic phenotyping is revolutionizing how we study tissue and immune responses to cancer treatments. In this talk, we will describe our integrated approaches to characterizing the tumour microenvironment in head and neck, lung, and skin cancers using ultra high-plex spatial proteomics and transcriptomics, benchmarking to ground truth, and developing functionally characterised immuno-metabolic signatures associated with resistance and sensitivity to immunotherapy.
Bio
Dr. Arutha Kulasinghe is a Senior Research Fellow and leads the Clinical-oMx Lab at the University of Queensland. Dr Kulasinghe has pioneered spatial transcriptomics using digital spatial profiling approaches in the Asia-Pacific region, contributing to world-first studies for lung cancer, head and neck cancer, skin cancer and COVID-19. His research aims to understand the underlying pathobiology by using an integrative multi-omics approach.
Cecilia Lindskog, Uppsala University, Sweden
Talk
A spatio-temporal single-cell type map of the human proteome based on transcriptomics, high-resolution antibody-based imaging and artificial intelligence
Abstract
For a fundamental understanding of human health, molecular medicine and targeted treatment, it is necessary to map processes unique to each tissue and cell type. We here aimed to set up a stringent, workflow for mapping human tissues at the single-cell type level, and utilized this workflow to create high-resolution spatio-temporal maps of tissue or cell type-specific proteins in human tissues. One of these tissues is testis, which is a complex organ with spermatogenesis involving thousands of genes and proteins activated or repressed through multiple cell states, from spermatogonial stem cells to mature sperm. Understanding the intricate functions and mechanisms at each step of this process requires a multi-dimensional approach that integrates both quantitative and qualitative methods.
Based on single-cell RNA sequencing data, we identified 12 distinct cell types and subsets of germ cells in testis, some of which cannot be distinguished by regular immunohistochemistry. Using a multiplex immunofluorescence pipeline, we then built antibody panels specifically outlining each of these cell types. The fixed antibody panels were stained together with the candidate protein of interest, one at a time, to pinpoint the exact protein localization at a cellular and subcellular level. Using artificial intelligence, the high-resolution images were quantified using an automated image analysis workflow. The integrated data allowed us to study temporal mRNA and protein expression gradients along with maturation processes and identify which mRNAs that are consistently translated into proteins from those that vary from a spatio-temporal aspect. We were also able to assign presumed functions to numerous uncharacterized proteins not previously described in the context of human reproduction.
In summary, we present a strategy for high-resolution spatio-temporal mapping of human tissues and cells, presented as a single-cell type reference map of adult human testis. The data which is freely available on www.proteinatlas.org decodes the complexity of human germ cells and links quantitative data with tissue morphology, and the validated workflow has the potential to be used for other tissues, contributing to valuable insights into molecular function and processes linked to disease.
Bio
Dr. Lindskog is a research group leader and associate professor at the Department of Immunology, Genetics and Pathology, Cancer Precision Medicine unit, Uppsala University, Sweden. Her research group uses a multi-dimensional approach and combination of various qualitative and quantitative methods to generate high-resolution spatial maps of the human body at the single-cell type level. By combining scRNA-seq, spatial technologies for mRNA and protein detection and machine learning, she aims to link single cell type-specific expression profiles with molecular function and mechanisms of disease, a first step towards precision medicine. Since 2006, Dr. Lindskog is also affiliated with the Human Protein Atlas (HPA) project, the largest biological database for spatial proteomics, publicly available at www.proteinatlas.org. She received her PhD in Pathology at Uppsala University in 2013, and since 2014 she has been director of the tissue-based efforts of the HPA project, leading the team that generates the mRNA and protein expression data in human tissues.
Joakim Lundeberg, KTH, Sweden
Talk
Tissue ecosystems in time and space
Abstract
Tools in spatial biology offer a wide range of technologies that quantify different types of biomolecules in tissue sections. These technologies provide information about distinct aspects of tissue anatomy, such as its morphology, genome, transcriptome, proteome, and metabolome. Here, several multiomics computational and experimental methodologies approaches that capture the tissue ecosystem from a single tissue section will be described.
Bio
Joakim Lundeberg, Ph.D., Professor in Molecular Biotechnology, has, during the most recent years, focused on spatial transcriptomics, which enables a detailed description of gene expression patterns in tissue sections. The methodology is now available worldwide through 10x Genomics Inc, as Visium. The technology was also featured in Nature Methods as the Method of the Year 2020. Dr. Lundeberg has publications demonstrating technology development into several new spatial modalities and examples of its impact in biology. The current research focuses on expanding the spatial modalities and developing new tools and applications in human cell atlas, neurology, and cancer.
Sinem Saka, EMBL, Germany
Talk
A DNA toolbox for spatial biology from imaging to sequencing
Abstract
DNA is not only a fundamental constituent of cells, but has also great capacity for information storage. We leverage the predictability of DNA hybridization kinetics and orthogonality of DNA sequences to utilize DNA oligos as tagging and barcoding tools for improving the major limitations of in situ visualization of molecules. We have previously developed multiplexed imaging approaches such as SABER-FISH and Immuno-SABER that utilize DNA barcoding and a flexible in situ signal amplification system for efficient visualisation of many protein, DNA or RNA targets in cells and tissues. More recently, we have implemented DNA barcoding for a new spatial transcriptomics approach, Light-Seq, which directly integrates fluorescence imaging and whole-transcriptome next-generation sequencing of the same cells in fixed biological samples. Light-Seq combines spatially-targeted, rapid photocrosslinking of DNA barcodes onto cDNAs in situ with a novel one-step DNA stitching reaction to create pooled, spatially-indexed sequencing libraries. This light-directed barcoding enables imaging-based in situ selection of multiple cell populations in intact fixed tissue samples for full transcriptome sequencing based on location, morphology, or protein stains, without cellular dissociation. Applying Light-Seq to mouse retinal sections, we discovered new biomarkers for a very rare neuronal subtype, dopaminergic amacrine cells, from only 4-8 individual cells per section. Light-Seq provides an accessible workflow to combine in situ imaging and protein staining with next-generation sequencing of the same cells, leaving the sample intact for further analysis post-sequencing. We are using these methods to directly link multi-dimensional and high-resolution microscopic phenotypes to transcriptomic profiles for diverse sample types.
Bio
PhD in Molecular Biology, 2013, University of Göttingen/International Max Planck Research School, Germany.
Postdoctoral research at Wyss Institute for Biologically Inspired Engineering, Harvard University, USA.
EMBO Postdoctoral Fellow 2016–2017.
Human Frontier Science Program Fellow 2017–2020.
Group leader at EMBL since December 2020.
Jeffrey Spraggins, Vanderbilt University, Nashville, USA
Talk
Bringing human disease into focus through integrated multimodal molecular imaging
Abstract
Cellular interactions within tissue micro-environments form the basis of health and disease for all organisms. Exposure to nutrients, toxins, and neighboring cells triggers coordinated molecular responses that impact cellular function and metabolism in a beneficial, adaptive, or detrimental manner. Acquiring molecular information at cellular resolution is thus crucial for developing a comprehensive understanding of the biology of an organism. Matrix-assisted laser desorption/ionization (MALD) imaging mass spectrometry (IMS) addresses this need by combining the spatial fidelity of classical microscopy with the molecular specificity of a mass spectrometer. This presentation will highlight our work developing new, high-performance technologies for improving the spatial resolution, sensitivity, and specificity of MALDI IMS for metabolite, lipid, and protein mapping. This will include the utilization of novel MALDI methods and development efforts using high spatial resolution Q-TOF platforms to address the molecular complexity associated with direct tissue analysis. Further, I will describe recent advances in integrated, multimodal methods that correlate molecular signals to specific biological tissue features and cell types. These technologies will be demonstrated through various biomedical research applications that include the construction of molecular atlases and understanding the molecular drivers of normal aging and disease.
Bio
Jeff Spraggins is an Associate Professor in the Department of Cell and Developmental Biology, Director of the Biomolecular Multimodal Imaging Center (BIOMIC), and Director of the Mass Spectrometry Research Center at Vanderbilt University. He received his B.A. in Chemistry from the College of Wooster and his Ph.D. in Analytical Chemistry from the University of Delaware. Dr. Spraggins’ research program focuses on the development of next-generation imaging mass spectrometry (IMS) technologies to elucidate the molecular basis of health and disease. Modern instrumentation and computing capabilities have enabled researchers to move beyond reductionist biology and, instead, probe how the components of biological entities (e.g., molecules, cells, and tissues) interact to reveal the underlying biology of disease. This systems biology approach has been accelerated by advancements in high-throughput ‘omics’ technologies; however, genetic and molecular information is only part of the story. The challenge lies in understanding how these parts interact and how perturbations to the system relate to disease. Molecular imaging effectively offers a ‘blueprint’ as to how biological components work together by providing spatial context to molecular information. Broadly, Dr. Spraggins’ research falls into two categories: (1) Developing novel mass spectrometry technologies to maximize imaging performance, enabling molecular histology at cellular resolution, and (2) combining imaging mass spectrometry with a variety of other biomedical imaging technologies to create new integrated modalities capable of providing a systems biology view of tissue at cellular resolution. The Spraggins group applies these technologies to the construction of molecular atlases as part of the NIH Human Biomolecular Atlas Program (HuBMAP), the NIDDK Kidney Precision Medicine Project (KPMP), and the NCI Human Tumor Atlas Network (HTAN); and also, to understanding the molecular drivers of Alzheimer’s, kidney, and infectious diseases.
Carolina Wählby, Uppsala University, Sweden
Talk
Measuring tissue morphology in spatial biology
Abstract
Spatial omics has transformed our understanding of tissue architecture by preserving spatial context of gene expression patterns. Simultaneously, advances in imaging AI have enabled extraction of morphological features describing the tissue. The intersection of spatial omics and imaging AI presents opportunities for a more holistic understanding: morphological features can be translated or integrated into spatial omics analyses. By translation we mean finding morphological features that spatially correlate with gene expression patterns with the purpose of predicting gene expression. Such features can be used to generate super-resolution gene expression maps or infer genetic information from clinical H&E-stained samples. By integration we mean finding morphological features that spatially complement gene expression patterns with the purpose of enriching information. Such features can be used to define spatial domains, especially where gene expression has preceded morphological changes and where morphology remains after gene expression.
Bio
Carolina Wählby is professor in Quantitative Microscopy at the Dept. of Information Technology, Uppsala University, and Scientific Director of the National SciLifeLab Bioimage Informatics facility. Her research lies in the intersection between life science and computational image analysis, developing image analysis, deep learning, and visualization approaches for understanding the organization and dynamics of cells and tissue, funded primarily by the ERC and the Swedish Foundation for Strategic research. She has a MSc in molecular biotechnology and a PhD in digital image analysis, and carried out postdoc research within genetics and pathology. She was part of the Imaging Platform of the Broad Institute 2009-2015, developing CellProfiler, and became full professor at Uppsala University in 2014, and is a member of the Royal Swedish Academy of Engineering Sciences.
Important dates
October 1: Science Summit Opens at Uppsala Konsert & Kongress
Exhibitors
Meet representatives from companies providing products and services in relevant fields. The following companies will be present during the day
- 10x Genomics
- Akoya Biosciences
- BioNordika AB
- Bruker (Canopy Biosciences and NanoStrings)
- Lunaphore
- Navinci Diagnostics AB
- Saveen & Werner AB
- Thermo Fisher Scientific
- Vizgen Incorporated
Scientific Committee
Mats Nilsson, Chair and Platform Director Spatial Biology, SciLifeLab/Stockholm University
Charlotte Stadler, Platform Co-Director Spatial Biology, SciLifeLab/KTH
Per Andrén, SciLifeLab/Uppsala University
Stefania Giacomello, SciLifeLab/KTH
Project leader Operations Office: Erika Bergqvist Erkstam

