Adaptation to salinity across the bacterial tree of life: comparing genomes to unravel molecular mechanisms
A study led by Anders Andersson (SciLifeLab/KTH) identified aspects of adaptation to salinity common across the whole spectrum of bacterial biodiversity. The researchers based their findings on comparisons of genomes of related bacteria obtained directly from aquatic environments differing in salt concentrations. These results expand our understanding of how the functioning of bacteria depends on physicochemical conditions, and what happens to the microbes when the environment changes.
Salinity is the major factor determining which bacteria live where, and most aquatic environments can be divided into three biomes: freshwater, marine, and brackish (intermediate salinity levels, as in e.g. the Baltic or the Caspian Sea). These microorganisms make the foundation of the ecosystems in which they live, having indispensable roles in nutrient and organic matter flows. Therefore, the impact of salinity on microbial life is key to understanding how and why freshwater, brackish, and marine environments differ. Moreover, as climate change and other consequences of human actions lead to changes in salinity, to comprehend how ecosystems will be affected the knowledge about bacterial responses is essential.
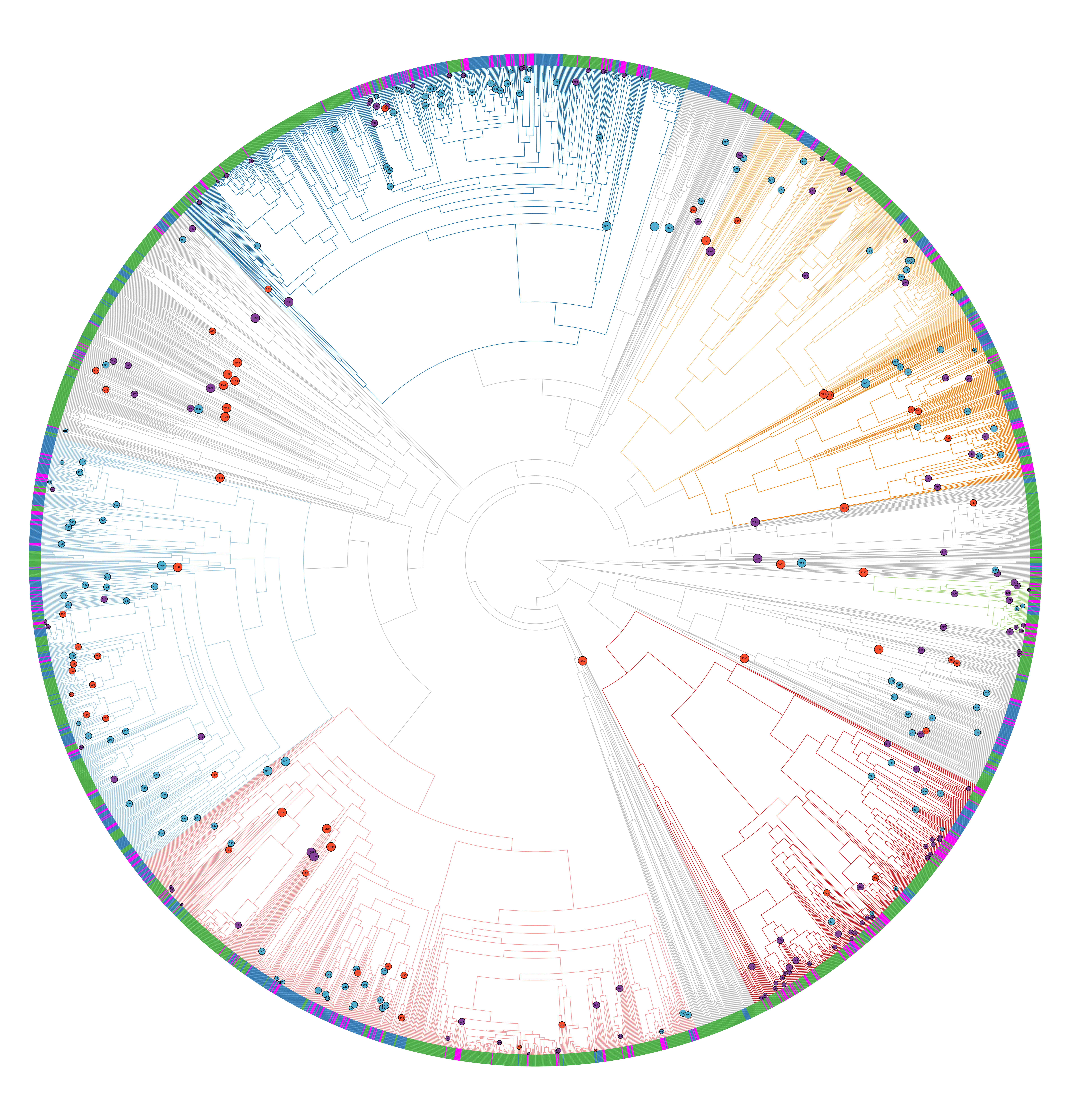
The authors of the study, Jurdzinski, Mehrshad, et al., found that almost all aquatic bacterial species can be found either only in freshwater, brackish, or marine environments. Transitions between these three biomes were rare, and almost all of them were estimated to have occurred at least millions of years ago, i.e. long before the Baltic Sea was born. The transitions were accompanied by consistent shifts in properties and amino-acid composition of the entire protein repertoire encoded by the genomes. These changes had undergone over tens of millions of years after bacteria switched to a different environment. Thus, proteome incompatibility is likely a major barrier separating bacteria between the biomes, which takes a long time to overcome. Furthermore, the same gene functions were discovered to be gained or lost in diverse groups of bacteria transitioning between the same biomes. These gene gains/losses point to a widespread mechanism of adaptation allowing transitions across salinity barriers, as well as improve our understanding of functioning and biotechnological potential of the identified genes.
The most likely responses of bacteria to changes in salinity can be inferred from these results. Considering the timescale of cross-biome transitions, local adaptation to abrupt change is unlikely. Indeed, researchers found that as big geological events as formations of the Baltic and the Caspian Seas did not result in more transitions then in other periods. Most likely, as salinity changes across the biome barriers, local bacterial populations die and the ecosystems are colonized by migrating bacteria. However, while salinity separated bacterial species, in the brackish biome within-species diversity was impacted more by geographic barriers than substantial differences in salt concentration. Thus, local salinity changes will likely lead to losses of the diversity contained within local populations and homogenization of bacterial species. It remains unknown how long it takes for bacteria to colonize an altered environment, and thus how long the ecosystems would remain deprived of their microbial foundations.