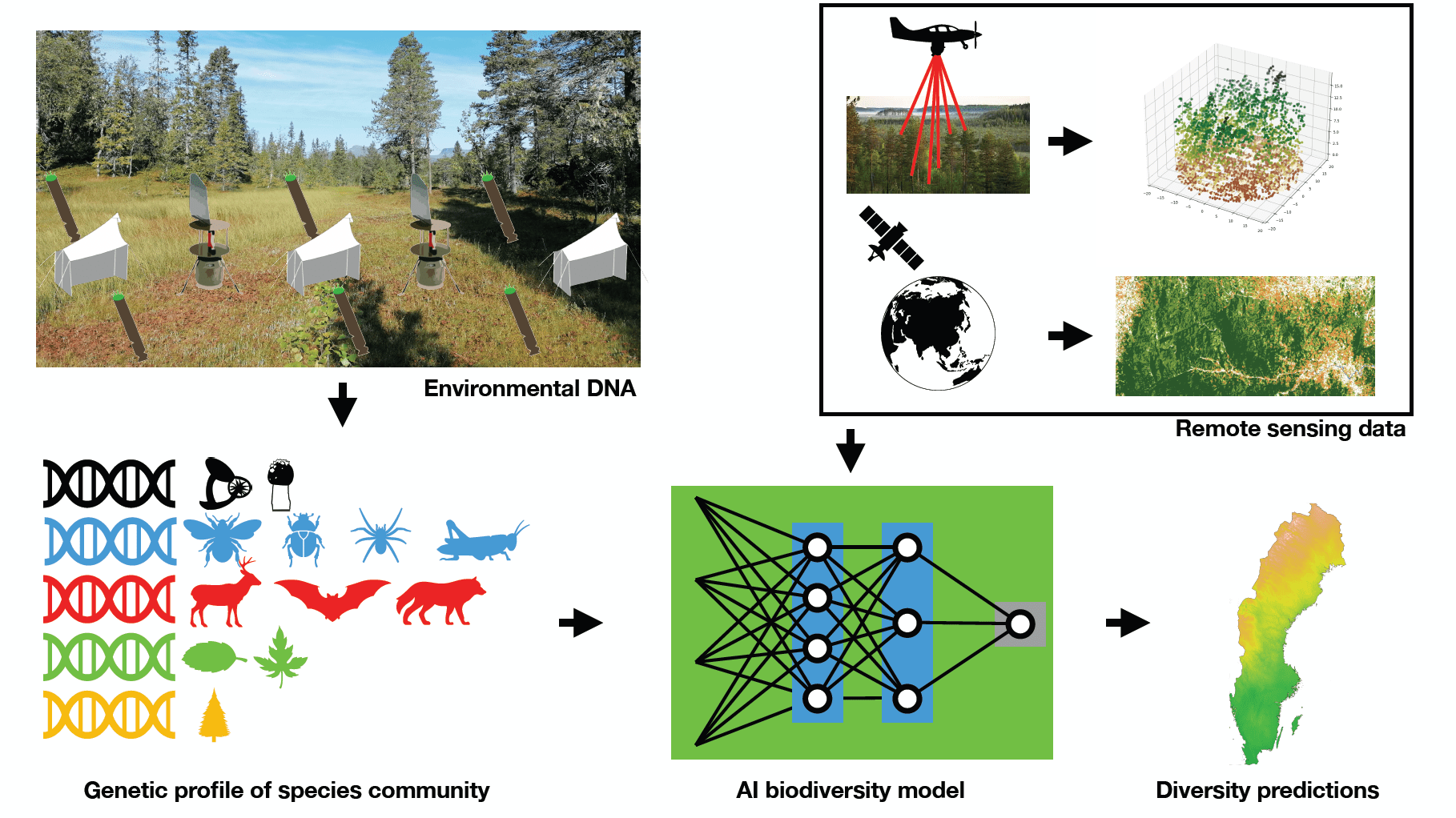
We are currently finding ourselves in the middle of an ongoing global biodiversity crisis. The formulation and execution of concrete national and international biodiversity goals and policies are hampered in great part by the fact that biodiversity is very complex and is not easily defined and quantified. Similar to concretely definable carbon-emission thresholds and offsets, we need to develop biodiversity metrics that in some way capture the complexity of lifeforms at a given site. We see this as one of the key challenges for the biological research community of our era, and we are dedicated to contributing to the development of such biodiversity metrics and measuring protocols through our research.
For this we are developing environment DNA sampling protocols and lab-processing techniques to reconstruct the diversity of a given species community across the whole tree of life. A particular focus lies on the often-neglected groups with staggering diversity, such as fungi, insects, or protists. We work in close collaboration with other labs with taxonomic and genetic expertise of these groups. With these environmental DNA samples and those collected by other researchers we contribute toward building a comprehensive database that allows us to detect the spatial distribution of taxa identified in our samples. We apply machine learning models to learn from these data and model biodiversity based on a range of biotic and abiotic predictors, including satellite images and airborne laser-scan data.
Group Members
Tobias Andermann (PI)
Adrian Baggström (PhD student)
Monica Guilera Recoder (PhD student)
Mirjam Lichtner (Master student)
